This page includes a narrative overview of my research, organized into different ‘themes’.
You can also skip right to my publications.
Research Overview
The human brain is constantly receiving dizzying amounts of information. Despite this, it is (usually) able to select important inputs, process what they mean, and come up with appropriate responses. Doing all this requires powerful processes to parse, select, and organize incoming information, and flexibly choose and direct systematic outputs. How is it that the brain is able to organize all of its seemingly chaotic activity in order to allow us to systematically respond in structured ways, ultimately allowing for allowing us to do everything that we do? In my research, I try to understand how patterns of neural activity contribute to this high degree of behavioral flexibility, in which we are able to quickly and flexibly react to an ever changing world.
Alternately put, the brain must have some powerful organizational capacities for coordinating activity, allowing for the functional organization of brain activity. With this idea in mind, in my work I focus on investigating patterns of electrical brain activity that arise from large groups of neurons. In particular, I’m involved in work focused on measuring, analyzing and interpreting both periodic activity, or neural oscillations, and aperiodic activity, sometimes described as ‘1/f activity’, that we see in electrical recordings of the brain. From these signals we try and infer properties of physiological activity, and how this activity may relate to cognition and disease. This work includes methods development of software tools, analyzing data from open-access databases, and running novel experiments with human subjects while we record their brain activity.
In practice, my research falls into a few different components, including:
- 1) Methods Development: developing methods and software tools to analyze neural data
- 2) Descriptive Work: exploring properties and patterns of neural data at scale
- 3) Experimental Work: hypothesis-driven experiments of periodic & aperiodic neural activity
- 4) Physiological Work: examining the activity of individual neurons
- 5) Text-Mining & Meta-Science: automated meta-analyses on the scientific literature
Most of the projects currently described were done as part of my PhD thesis at the Voytek Lab, with Professor Bradley Voytek, in the department of Cognitive Science at the University of California, San Diego.
Projects from my current postdoctoral work in the Jacobs Lab, in the Biomedical Engineering department at Columbia University will be added soon.
On the rest of this page, you can find descriptions of the following projects:
- Methods Development: Measuring Periodic & Aperiodic Activity
- Descriptive Work: Electrophysiological Data Mapping
- Experimental Work: Task-Related Periodic & Aperiodic Activity
- Physiological Work: Examining the Activity of Individual Neurons
- Meta-Science: Automated Meta-Analyses of the Scientific Literature
Methods Development: Measuring Periodic & Aperiodic Activity
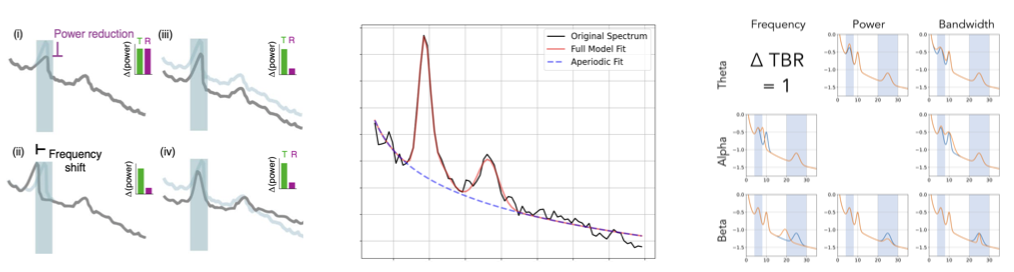
Quick Version: Methods for measuring periodic and aperiodic neural activity.
Neuro-electrophysiological data contains periodic or rhythmic components, often referred to as ‘neural oscillations’, as well as an aperiodic or ‘1/f-like’ activity. These different periodic & aperiodic components have different physiological generators and interpretations, such that we argue that they should each be explicitly measured in order to ensure that analyses and interpretations appropriately describe which features of the data are changing. To do so, we have developed a method to parameterize these components from the neural power spectrum. The method for parameterizing neural power spectra is available as an open-source Python toolbox, and is described in this paper.
In related work, we evaluate and compare other methods, including band-ratio measures, such as theta/beta, which systematically conflate periodic and aperiodic activity, and methods for analyzing aperiodic activity. More generally, we created the “Oscillation Methods” project, a ‘methodological review’ of considerations for measuring and interpreting neural oscillations, which is described in this paper and also hosted on this project website. These methods-related projects are largely driven by simulated data, including with our NeuroDSP toolbox, which is described in this paper.
Descriptive Work: Electrophysiological Data Mapping
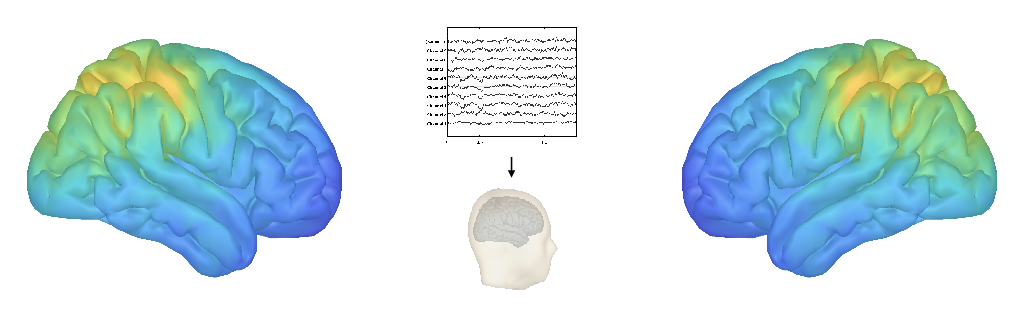
Quick Version: The ‘what, when, and where’ of neuro-electrophysiological data.
Descriptively, I work on projects investigating the ‘where, when, and what’ of periodic and aperiodic neural activity, characterizing both periodic and aperiodic activity across the human cortex, and how this varies across people across different ages, and within people across different brain states.
In order to analyze and describe typical patterns of neural activity, I work on projects that analyze large, open-access datasets, and apply our novel methods to measure periodic and aperiodic components. This includes analyzing a large datasets of magnetoencephalography (MEG) data (included in the parameterization paper) and EEG data (included in the band-ratios paper). A follow-up report on this project is currently in preparation, with some additional analyses also currently available in conference posters (SfN2018, Biomag2018, Neuroinformatics2018).
Beyond examining patterns of periodic & aperiodic activity in healthy adult populations, I am also involved in projects investigating neural activity across development and in different brain states. In developmental work, this includes work examining systematic changes in aperiodic activity across childhood (work led by Wei He, presented in this preprint, as well as a tutorial paper for applying spectral parameterization to developmental data (work led by Brendan Ostlund, presented in this paper. Work examining patterns of activity across different brain states includes analyses of aperiodic activity during anesthesia (including in this paper), and across sleep stages (work led by Mohamed S. Ameen, presented in this preprint.
In addition, I contribute to clinical work, across multiple disease contexts. For example, in a project led by Madeline Robertson, we analyzed EEG data from children with ADHD, and find that there is a systematic different in aperiodic activity in this group, as described in this paper. In another project, led by Martina Kopčanová, we analyzed data from patients with Alzheimer’s dementia, demonstrating that in this case, it is periodic, but not aperiodic activity that differentiates clinical from control populations, as presented in this paper.
Experimental Work: Task-Related Periodic & Aperiodic Activity
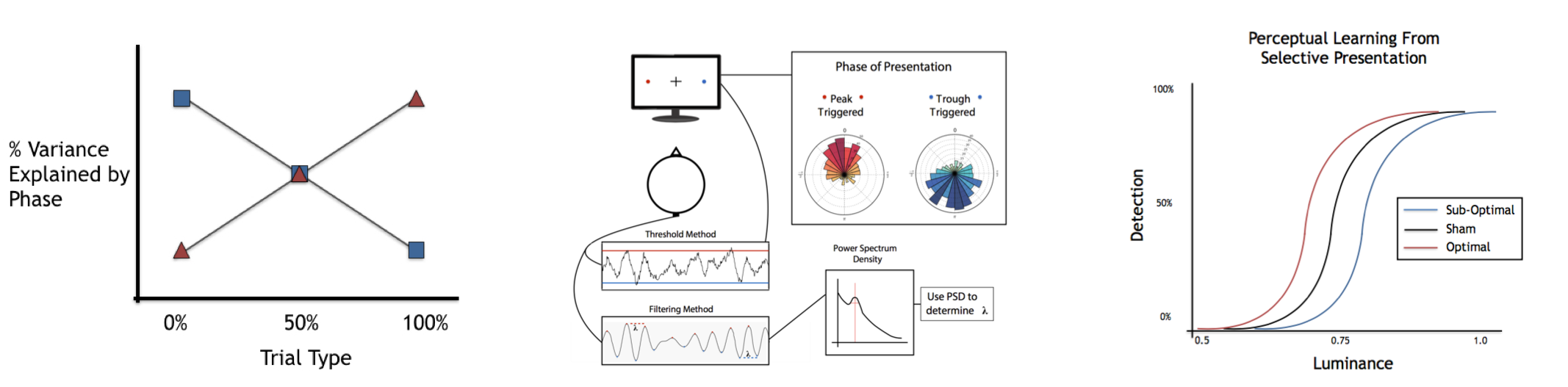
Quick Version: How do momentary fluctuations in neural activity relate to cognition?
If I repeatedly flash a very brief, dim light, for the exact same event that happens, sometimes you may see it, but sometimes you won’t. Why does the brain react so differently to the same input? One finding is that oscillatory phase, that is, the precise part of the rhythmic brain activity at which the flash occurs, systematically relates to your perception. The underlying idea is here is that low-frequency oscillations may work to parse incoming inputs - this selection process creating rhythmic fluctuations of better (and worse) processing. To investigate these ideas, we have developed an online presentation system that can selectively present stimuli at particular phases of subject’s ongoing oscillations that we can use to investigate these ideas. A preprint of this project is upcoming. The online presentation brain-computer interface and task validation was presented at SfN-2015. A follow up (offline) experiment examining the role of oscillatory phase in perceptual and/or attentional processes was presented at SfN-2016.
In other work, we investigate task-related dynamics of aperiodic neural activity. In work led by Leo Waschke, we did an experiment in which we presented subjects with stimuli that itself has 1/f properties. By manipulating both the stimulus characteristics, and the attentional focus of the subjects, we were able to show how perceptual and attentional task elements systematically affect event-related aperiodic neural activity. This work is described in this paper.
Physiological Work: Examining the Activity of Individual Neurons
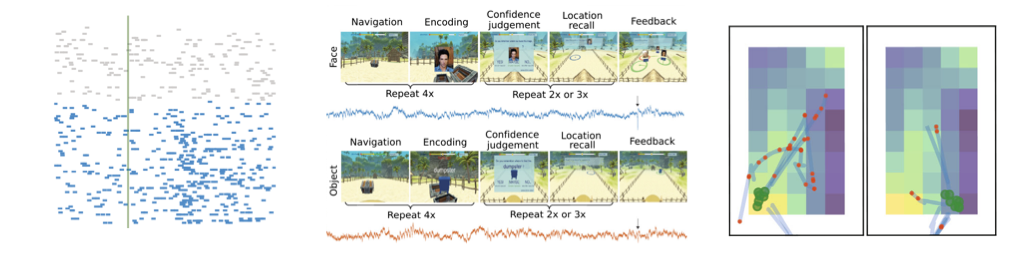
Quick Version: Investigating patterns of single-unit activity.
The aforementioned work examines patterns of neural activity that can be examined from the local field potential, reflecting the contribution of thousands to millions of individual neurons. In order to better understand the activity of individual neurons (and ultimately, how this contributes to the local field), I also have a line of research in which I analyze the activity of individual neurons that can be recorded from awake, behaving humans who are in the hospital for epilepsy monitoring.
Methodologically, my work in single-units includes developing the Human Single Unit pipeline, a pipeline for managing and doing projects with single-unit activity including the use of standardized data formats as well as the [spiketools]https://github.com/spiketools/spiketools) Python module for analyzing single-unit recordings. Empirically, with Claire Z Han and Runnan Cao we did ran an experiment examining neural responses to faces and objects, comparing between an n-back memory task and a spatial navigation episodic memory task. In this work, we find neurons in the human medial temporal lobe that flexibly shift representations across different behavioral contexts, which is presented in this paper.
Meta-Science: Automated Meta-Analyses of the Scientific Literature
Quick Version: Computationally summarizing and investigating the scientific literature.
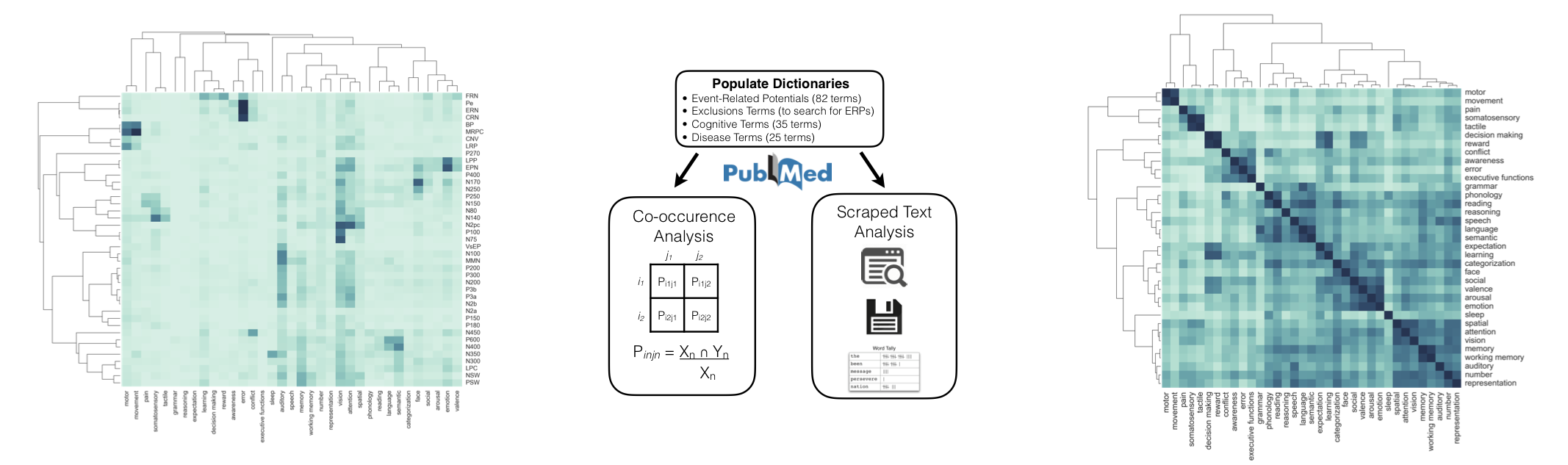
The scientific literature is vast and ever growing. While informatics offers tools and approaches to help parse, organize and summarize the literature, there has been relatively little adoption of these approaches in cognitive neuroscience. To address this, I work on tools and projects that aim to investigate and summarize patterns in the scientific literature. To do so, I created a Python module for performing collection and analysis of the scientific literature, which is available here, and described in this paper.
This tool, which uses the PubMed API to find and collect relevant literature, allows us to systematically and automatically analyze large collections of scientific literature. For example, we did an automated meta-analysis of studies using event-related potentials (ERPs), using automated procedures to create data-driven summaries of ERPs. This project is described in this paper, and hosted on a project website.
Working with Richard Gao, this work has been generalized to explore topics across cognition, resulting in a couple conference papers (Cogsci2017, CCN2019). In work led by Will Fox, we have extended this project to investigating aspects of scientific communication, in particular examining differences in conveyed confidence in the scientific literature, comparing between primary literature and press releases, which resulted in a conference paper.